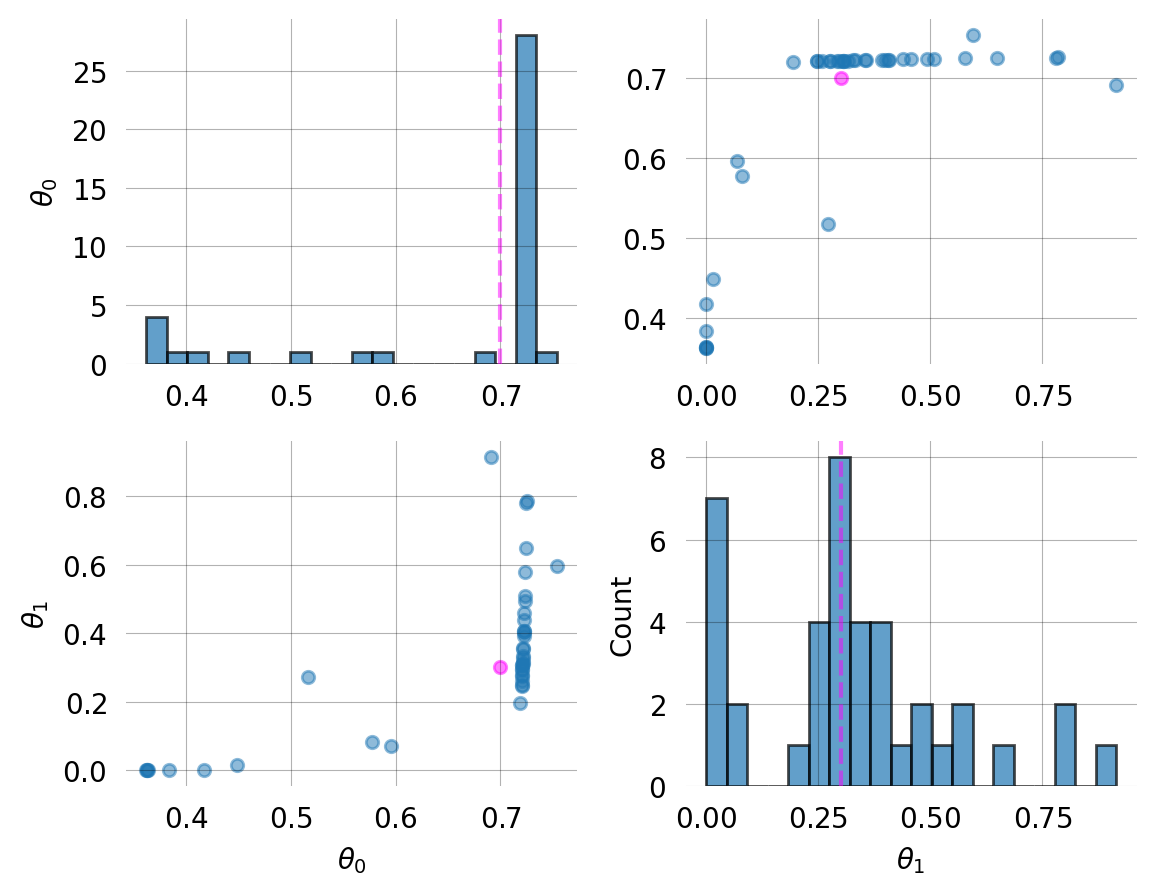
Lets try to estimate both coalescent and migration rate in a two population model.
from phasic import (# ALWAYS import phasic first to set jax backend correctly import numpy as npimport jax.numpy as jnpimport pandas as pdfrom typing import Optionalimport matplotlib.pyplot as pltfrom matplotlib.colors import LogNormimport seaborn as snsfrom tqdm.auto import tqdmfrom vscodenb import set_vscode_theme42 )'tab10' )'WARNING' )
Overriding theme from NOTEBOOK_THEME environment variable. <phasic._DeviceListFilter object at 0x14f9bd6d0>
<phasic.logging_config.set_log_level at 0x17f354490>
= 2 = StateIndexer(= ['pop1' , min_value= 0 , max_value= nr_samples),'pop2' , min_value= 0 , max_value= nr_samples),'in_pop' , min_value= 1 , max_value= 2 ),= [0 ] * indexer.state_length# set initial state with all lineages having one descendant at both loci = 1 , pop2= 0 , in_pop= 1 )] = nr_samples@with_ipv (initial)def coalescent_islands(state):= []if state[indexer.descendants.indices()].sum () <= 1 :return transitionsfor i in range (indexer.descendants.state_length):if state[i] == 0 : continue = indexer.descendants.index_to_props(i)for j in range (i, indexer.descendants.state_length):if state[j] == 0 : continue = indexer.descendants.index_to_props(j)if props_j.in_pop != props_i.in_pop:continue = int (i == j)if same and state[i] < 2 :continue if not same and (state[i] < 1 or state[j] < 1 ):continue = state.copy()-= 1 -= 1 = props_i.pop1 + props_j.pop1= props_i.pop2 + props_j.pop2if des_pop1 <= nr_samples and des_pop2 <= nr_samples:= indexer.descendants.props_to_index(= des_pop1, = des_pop2, = props_i.in_pop+= 1 * (state[j]- same)/ (1 + same), 0 ]])if state[i] > 0 := state.copy()= 2 if props_i.in_pop == 1 else 1 = state.copy()-= 1 = indexer.descendants.props_to_index(= props_i.pop1, = props_i.pop2, = other_pop+= 1 0 , state[i]]])return transitions= Graph(coalescent_islands) = [0.7 , 0.3 ]= 'LR' , by_index= lambda i: f"Desc in pop1: { indexer. index_to_props(i). descendants. pop1} " )
Overriding theme from NOTEBOOK_THEME environment variable. <phasic._DeviceListFilter object at 0x14f9bd6d0>
= 'LR' , by_state= lambda s: f"In pop1: { s[indexer.descendants.props_to_index(pop1= 1 )]. sum ()} " )
Overriding theme from NOTEBOOK_THEME environment variable. <phasic._DeviceListFilter object at 0x14f9bd6d0>
= graph.sample(1000 )
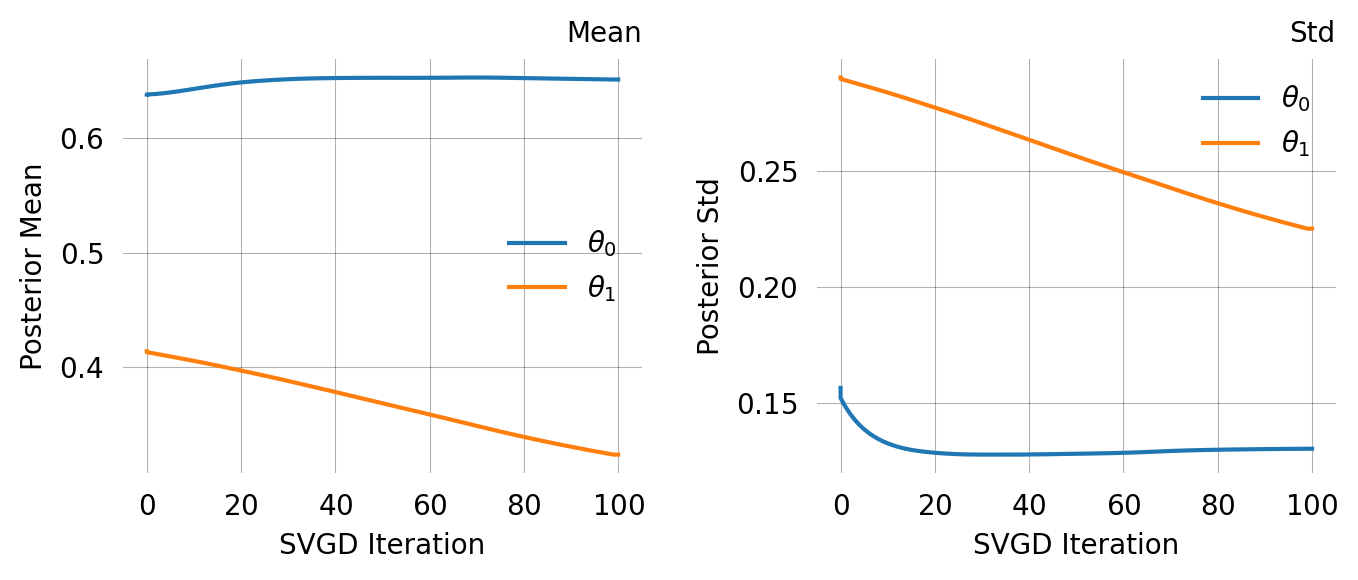
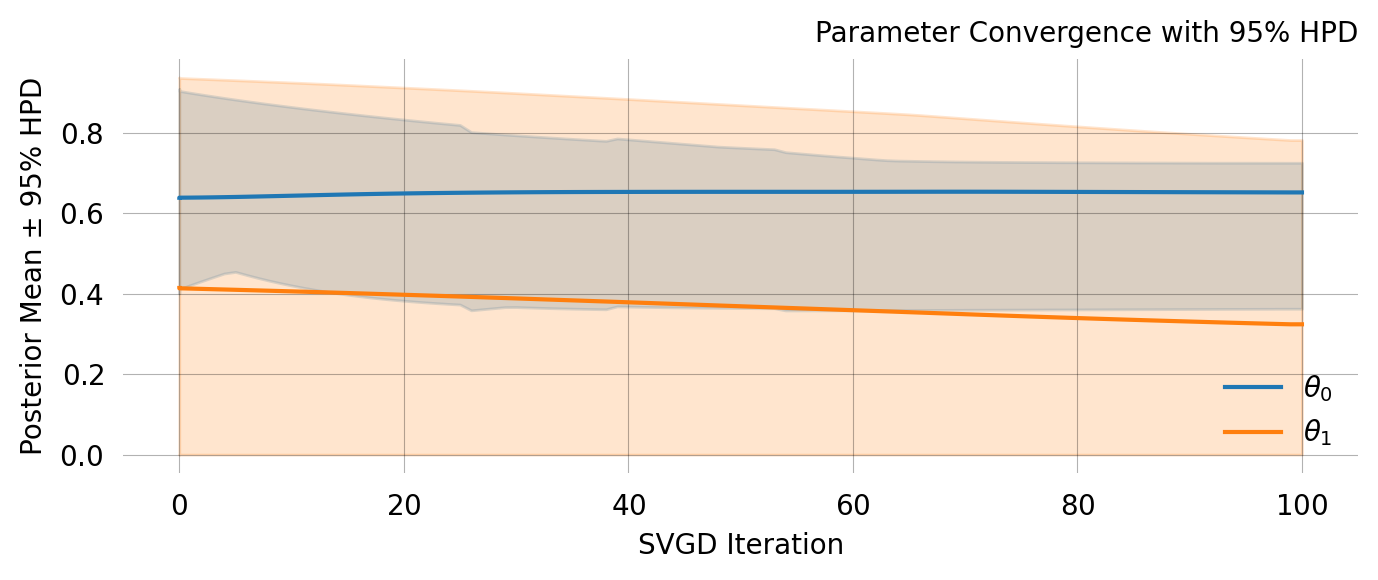
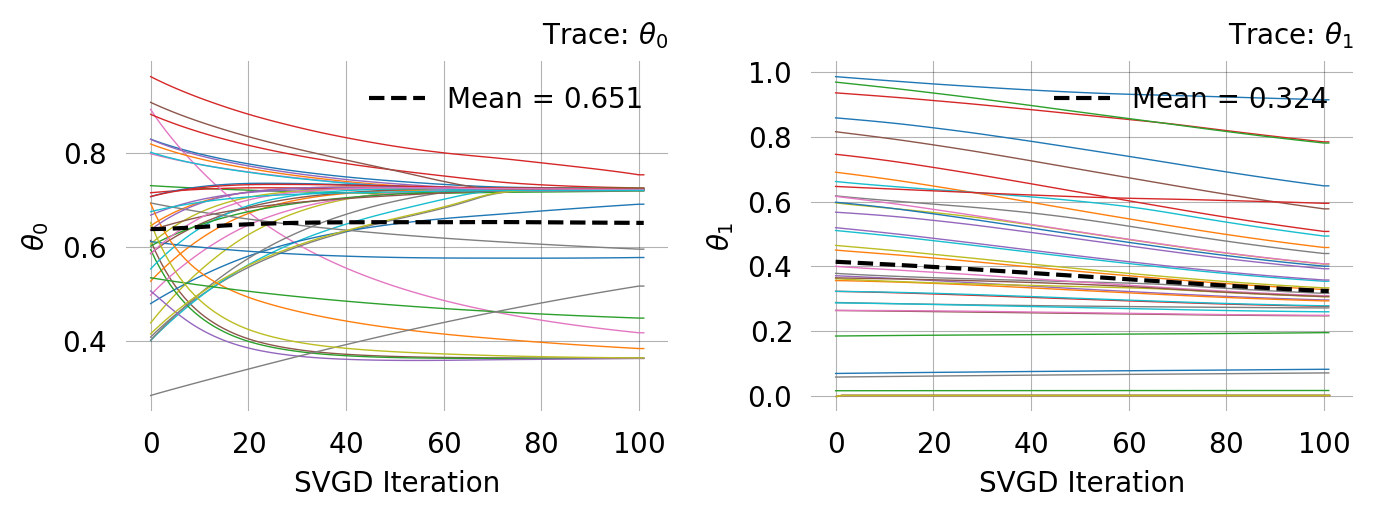
# quick run to get reasonable priors = graph.svgd(observations, n_particles= 5 , n_iterations= 20 )= svgd.get_results()= []for mean, std in zip (res['theta_mean' ], res['theta_std' ]):# full run with informed priors = graph.svgd(observations, prior= gauss_priors)
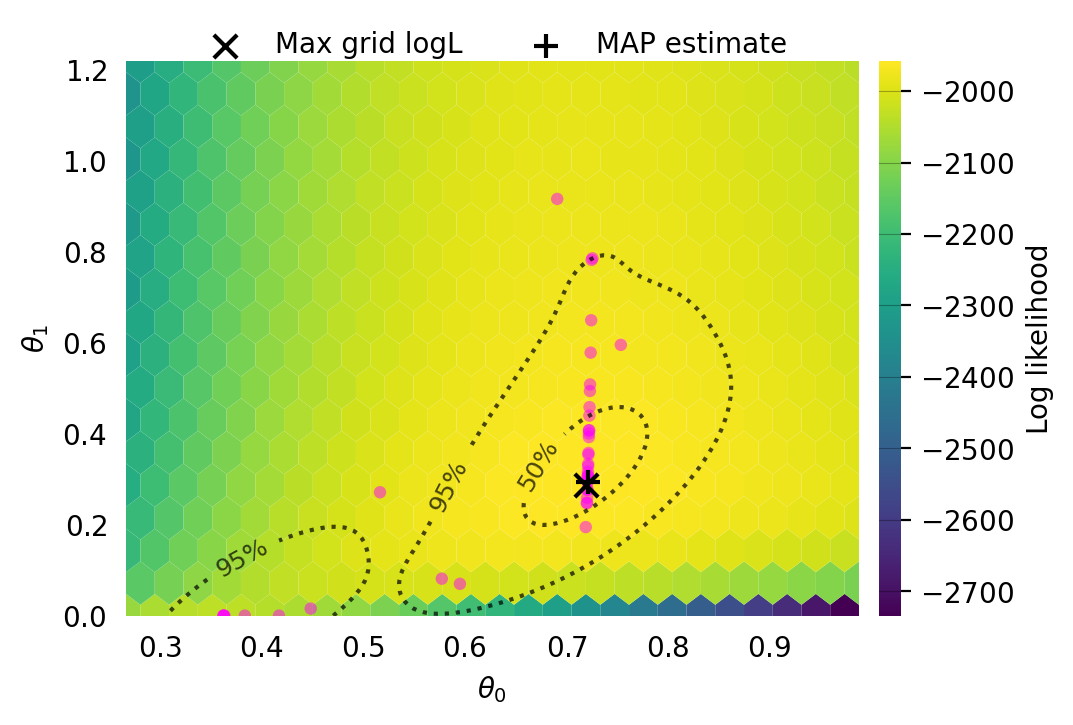
Parameter Fixed MAP Mean SD HPD 95% lo HPD 95% hi
0 No 0.7210 0.6512 0.1303 0.3622 0.7249
1 No 0.2929 0.3236 0.2250 0.0000 0.7813
Particles: 40, Iterations: 100
<Figure size 640x480 with 0 Axes>
= true_theta)
#svgd.animate_pairwise(true_theta=true_theta)