from phasic import (
Graph, with_ipv, GaussPrior, MoMResult, ProbMatchResult,
Adam, ExpStepSize, clear_caches,
StateIndexer, Property,
) # ALWAYS import phasic first to set jax backend correctly
import numpy as np
import jax.numpy as jnp
import matplotlib.pyplot as plt
import seaborn as sns
np.random.seed(42)
try:
from vscodenb import set_vscode_theme
set_vscode_theme()
except ImportError:
pass
sns.set_palette('tab10')Method of Moments
Data-informed priors via moment matching
A common challenge in Bayesian inference with SVGD is choosing sensible prior distributions. A vague prior may lead to slow convergence or poor exploration of the posterior, while an overly tight prior can bias the result. The method_of_moments method provides a principled way to construct data-informed priors by finding parameter estimates that match the model’s theoretical moments to the empirical moments of the observed data.
The method solves the nonlinear least-squares problem:
\hat{\theta}_{\text{MoM}} = \arg\min_{\theta > 0} \left\| \mathbf{m}(\theta) - \hat{\mathbf{m}} \right\|^2
where \mathbf{m}(\theta) are the model moments and \hat{\mathbf{m}} are the sample moments computed from the data. The standard error of the estimator is obtained via the delta method:
\text{Cov}(\hat\theta) \;=\; \left(\frac{\partial \mathbf{m}}{\partial \theta}\right)^{-1} \text{Cov}(\hat{\mathbf{m}}) \left(\frac{\partial \mathbf{m}}{\partial \theta}\right)^{-T}
where \text{Cov}(\hat{\mathbf{m}}) is estimated from the data. The point estimate and standard error are then used to construct Gaussian priors centred on the MoM estimate.
This is fast (seconds, not minutes) and gives SVGD a much better starting point than a generic prior.
Single-parameter model
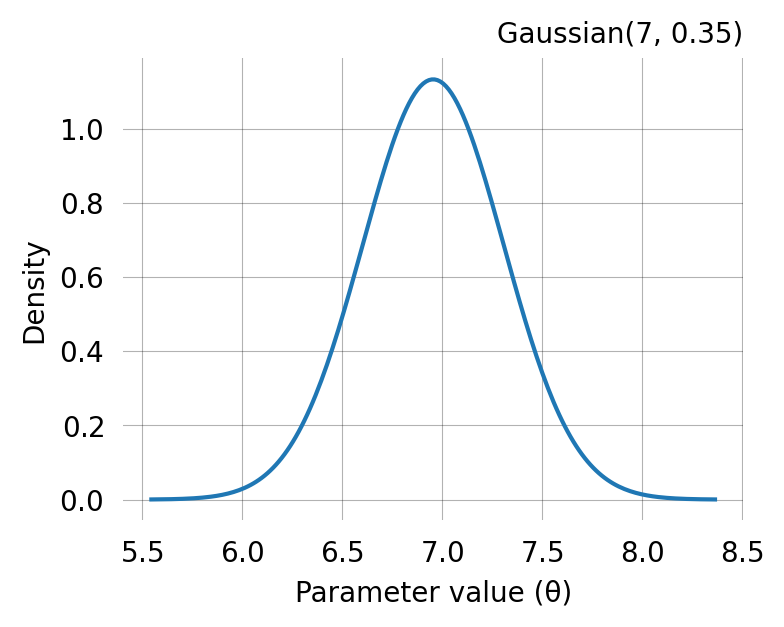
We start with the simplest case: a single-parameter exponential model. The coalescent rate \theta governs how quickly lineages coalesce. For an exponential distribution, the method of moments has an analytical solution: \hat{\theta} = 1 / \bar{x}.
nr_samples = 4
@with_ipv([nr_samples]+[0]*(nr_samples-1))
def coalescent_1param(state):
transitions = []
for i in range(state.size):
for j in range(i, state.size):
same = int(i == j)
if same and state[i] < 2:
continue
if not same and (state[i] < 1 or state[j] < 1):
continue
new = state.copy()
new[i] -= 1
new[j] -= 1
new[i+j+1] += 1
transitions.append([new, [state[i]*(state[j]-same)/(1+same)]])
return transitions
graph = Graph(coalescent_1param)
graph.plot()Sample observed data from the model with a known true parameter value:
true_theta = [7]
graph.update_weights(true_theta)
observed_data = graph.sample(1000)Run method of moments to find the parameter estimate:
mom = graph.method_of_moments(observed_data)MoM: theta_dim=1, n_free=1, nr_moments=4, n_features=1, n_equations=4
MoM: sample moments =
[0.21378016 0.07103006 0.03323121 0.01987239]
MoM: initial guess (full theta) = [6.95192796]
MoM: converged — `gtol` termination condition is satisfied.
MoM: theta = [6.95430573]
MoM: residual = 1.002189e-05
MoM: model moments =
[0.21569371 0.07007285 0.03146742 0.01834498]The result is a MoMResult dataclass containing the estimate, standard errors, and ready-to-use priors:
print(f"True theta: {true_theta}")
print(f"MoM estimate: {mom.theta}")
print(f"Std error: {mom.std}")
print(f"Converged: {mom.success}")
print(f"Residual: {mom.residual:.2e}")True theta: [7]
MoM estimate: [6.95430573]
Std error: [0.17612875]
Converged: True
Residual: 1.00e-05The MoMResult also reports how well the model moments match the sample moments:
print(f"Sample moments: {mom.sample_moments}")
print(f"Model moments: {mom.model_moments}")Sample moments: [0.21378016 0.07103006 0.03323121 0.01987239]
Model moments: [0.21569371 0.07007285 0.03146742 0.01834498]Using MoM estimates as SVGD priors
The main purpose of method_of_moments is to produce informed priors for SVGD. The mom.prior attribute is a list of GaussPrior objects that can be passed directly to graph.svgd():
svgd = graph.svgd(
observed_data,
prior=mom.prior,
optimizer=Adam(0.25),
)
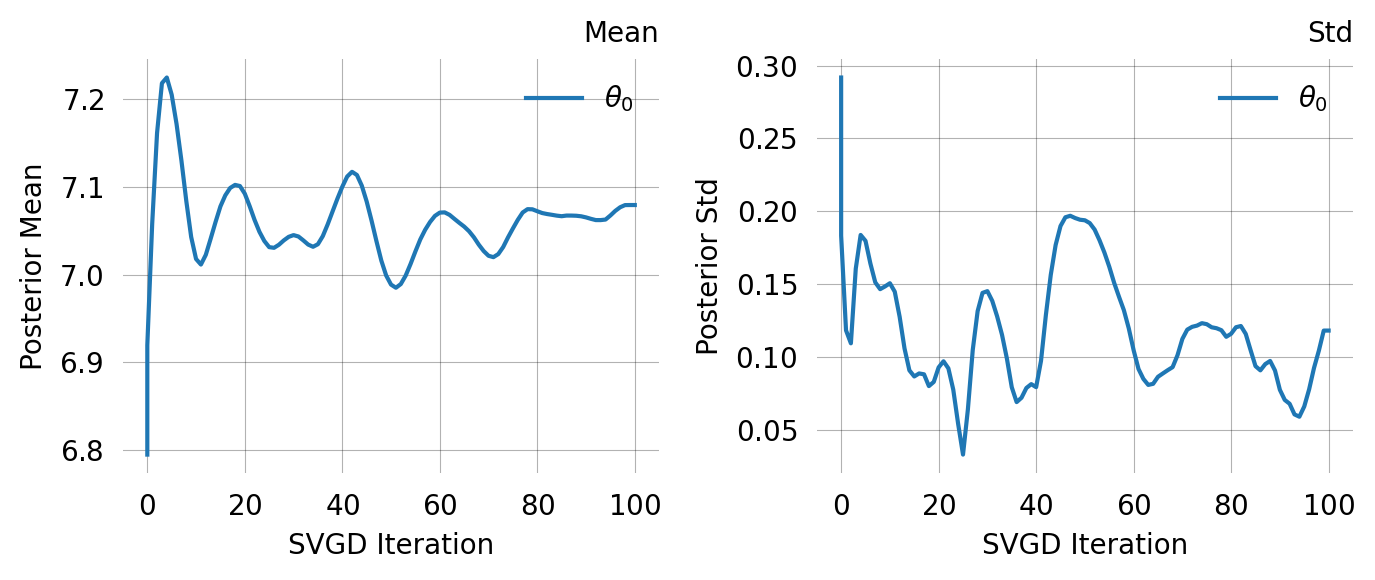
svgd.summary()
svgd.plot_convergence() ;Parameter Fixed MAP Mean SD HPD 95% lo HPD 95% hi
0 No 7.0606 7.0794 0.1180 6.7567 7.2409
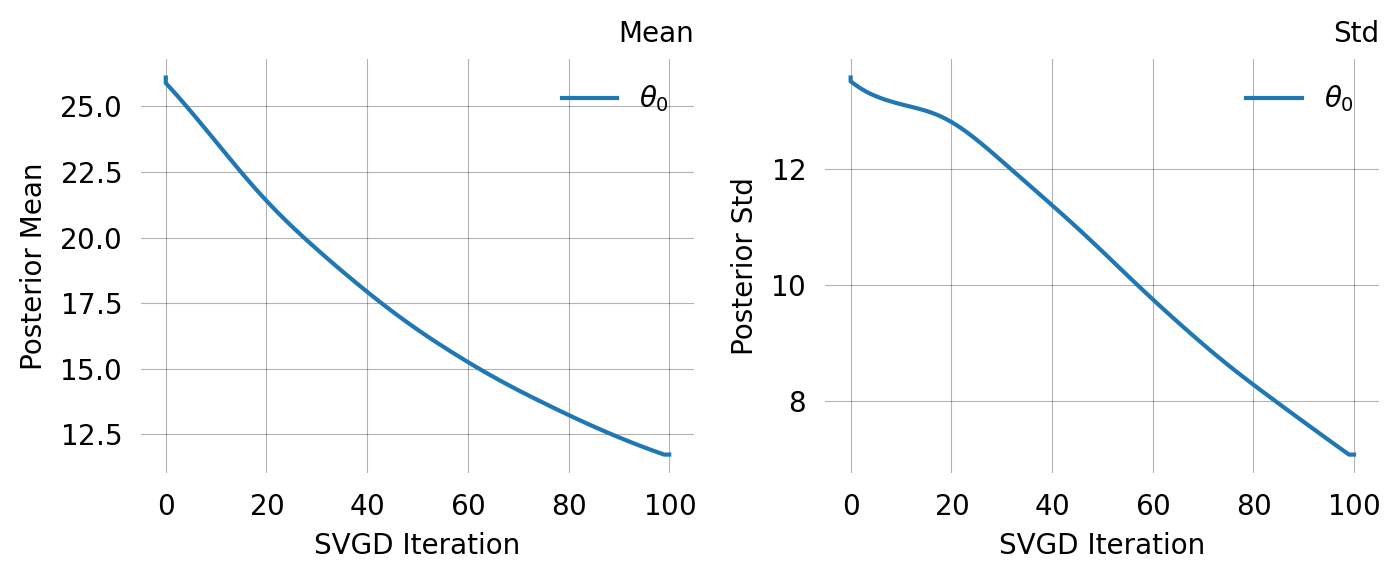
Particles: 24, Iterations: 100Compare this to using a vague prior. The MoM-informed prior starts SVGD in the right region, leading to faster and more reliable convergence:
Multi-parameter model
Method of moments is especially useful for multi-parameter models where choosing good priors by hand is difficult. Here we use a two-population island model with coalescent rate \theta_0 and migration rate \theta_1.
from phasic import StateIndexer, Property
nr_samples = 2
indexer = StateIndexer(
descendants=[
Property('pop1', min_value=0, max_value=nr_samples),
Property('pop2', min_value=0, max_value=nr_samples),
Property('in_pop', min_value=1, max_value=2),
])
initial = [0] * indexer.state_length
initial[indexer.descendants.props_to_index(pop1=1, pop2=0, in_pop=1)] = nr_samples
@with_ipv(initial)
def coalescent_islands(state):
transitions = []
if state[indexer.descendants.indices()].sum() <= 1:
return transitions
for i in range(indexer.descendants.state_length):
if state[i] == 0: continue
props_i = indexer.descendants.index_to_props(i)
for j in range(i, indexer.descendants.state_length):
if state[j] == 0: continue
props_j = indexer.descendants.index_to_props(j)
if props_j.in_pop != props_i.in_pop:
continue
same = int(i == j)
if same and state[i] < 2:
continue
if not same and (state[i] < 1 or state[j] < 1):
continue
child = state.copy()
child[i] -= 1
child[j] -= 1
des_pop1 = props_i.pop1 + props_j.pop1
des_pop2 = props_i.pop2 + props_j.pop2
if des_pop1 <= nr_samples and des_pop2 <= nr_samples:
k = indexer.descendants.props_to_index(
pop1=des_pop1, pop2=des_pop2, in_pop=props_i.in_pop)
child[k] += 1
transitions.append([child, [state[i]*(state[j]-same)/(1+same), 0]])
if state[i] > 0:
child = state.copy()
other_pop = 2 if props_i.in_pop == 1 else 1
child[i] -= 1
k = indexer.descendants.props_to_index(
pop1=props_i.pop1, pop2=props_i.pop2, in_pop=other_pop)
child[k] += 1
transitions.append([child, [0, state[i]]])
return transitions
graph = Graph(coalescent_islands)
graph.plot()true_theta = [0.7, 0.3]
graph.update_weights(true_theta)
observed_data = graph.sample(1000)Run method of moments on the two-parameter model:
mom = graph.method_of_moments(observed_data)MoM: theta_dim=2, n_free=2, nr_moments=4, n_features=1, n_equations=4
MoM: sample moments =
[2.89041844e+00 2.19034775e+01 2.64810856e+02 4.27038260e+03]
MoM: initial guess (full theta) = [ 0.66306831 12.49249157]
MoM: converged — Both `ftol` and `xtol` termination conditions are satisfied.
MoM: theta = [0.68703279 0.28364415]
MoM: residual = 3.242595e-02
MoM: model moments =
[2.91106921e+00 2.20801995e+01 2.64783140e+02 4.27038342e+03]print(f"True theta: {true_theta}")
print(f"MoM estimate: {mom.theta}")
print(f"Std error: {mom.std}")
print(f"Converged: {mom.success}")True theta: [0.7, 0.3]
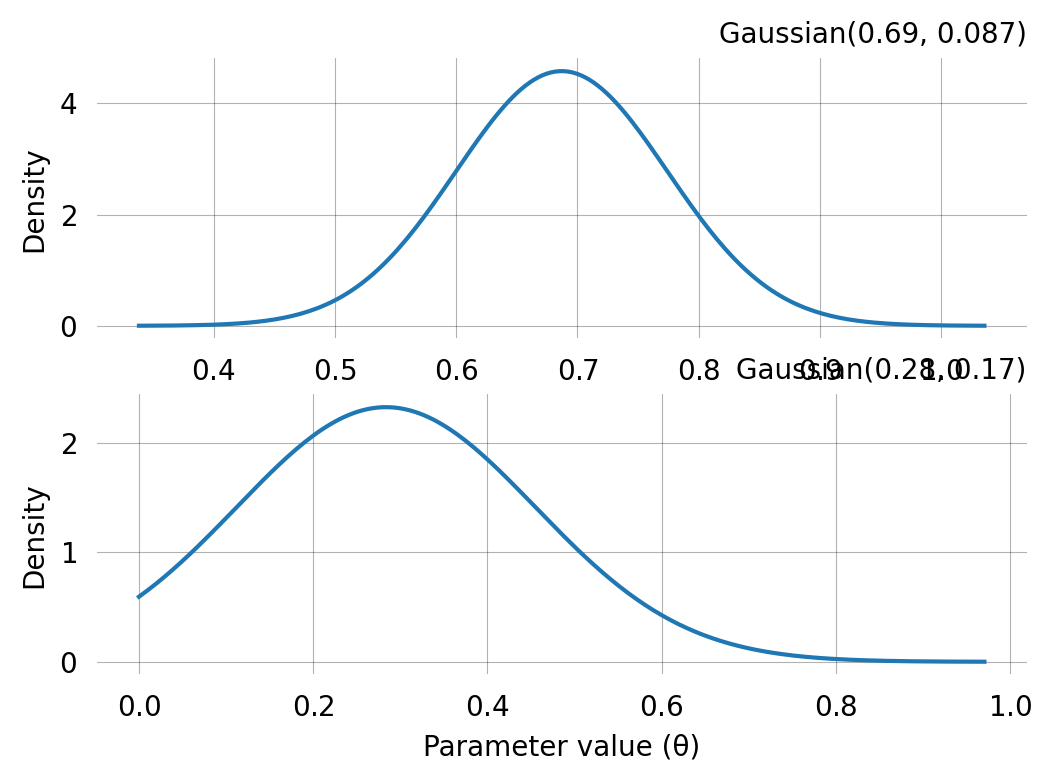
MoM estimate: [0.68703279 0.28364415]
Std error: [0.04358689 0.08585278]
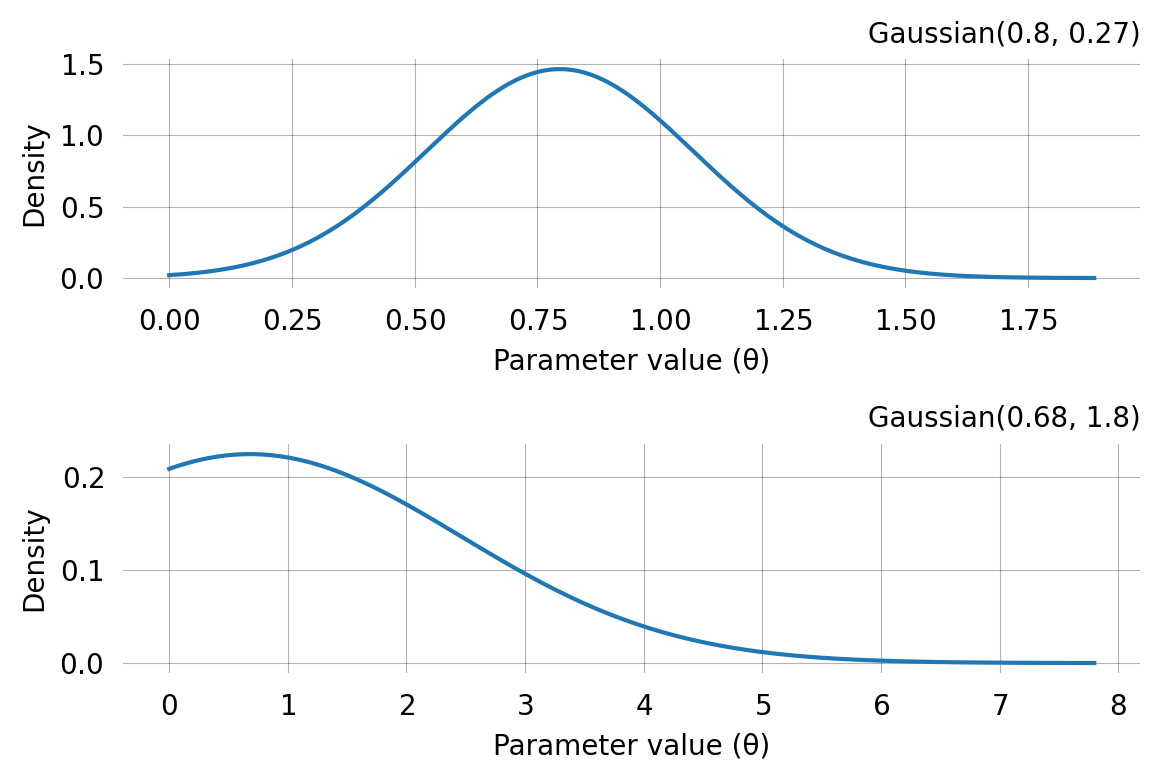
Converged: Truelen(mom.prior)2fig, axes = plt.subplots(len(mom.prior), 1, figsize=(6,4))
print(axes)
for i, prior in enumerate(mom.prior):
prior.plot(return_ax=True, ax=axes[i])[<Axes: > <Axes: >]Use the MoM priors for SVGD inference:
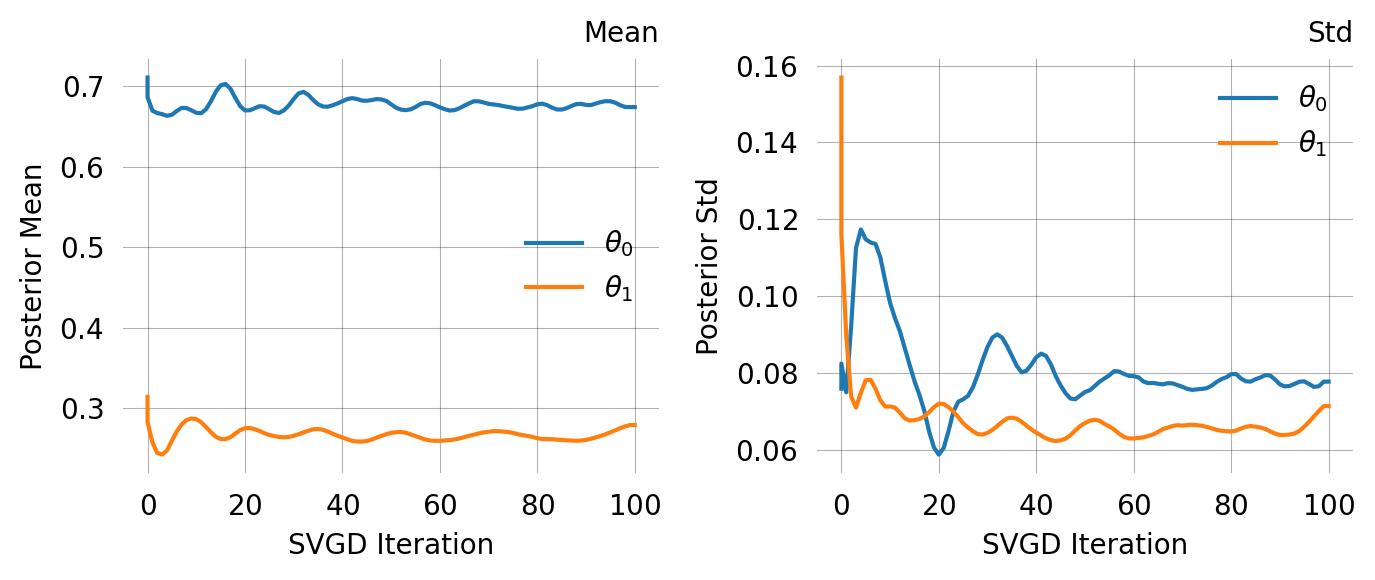
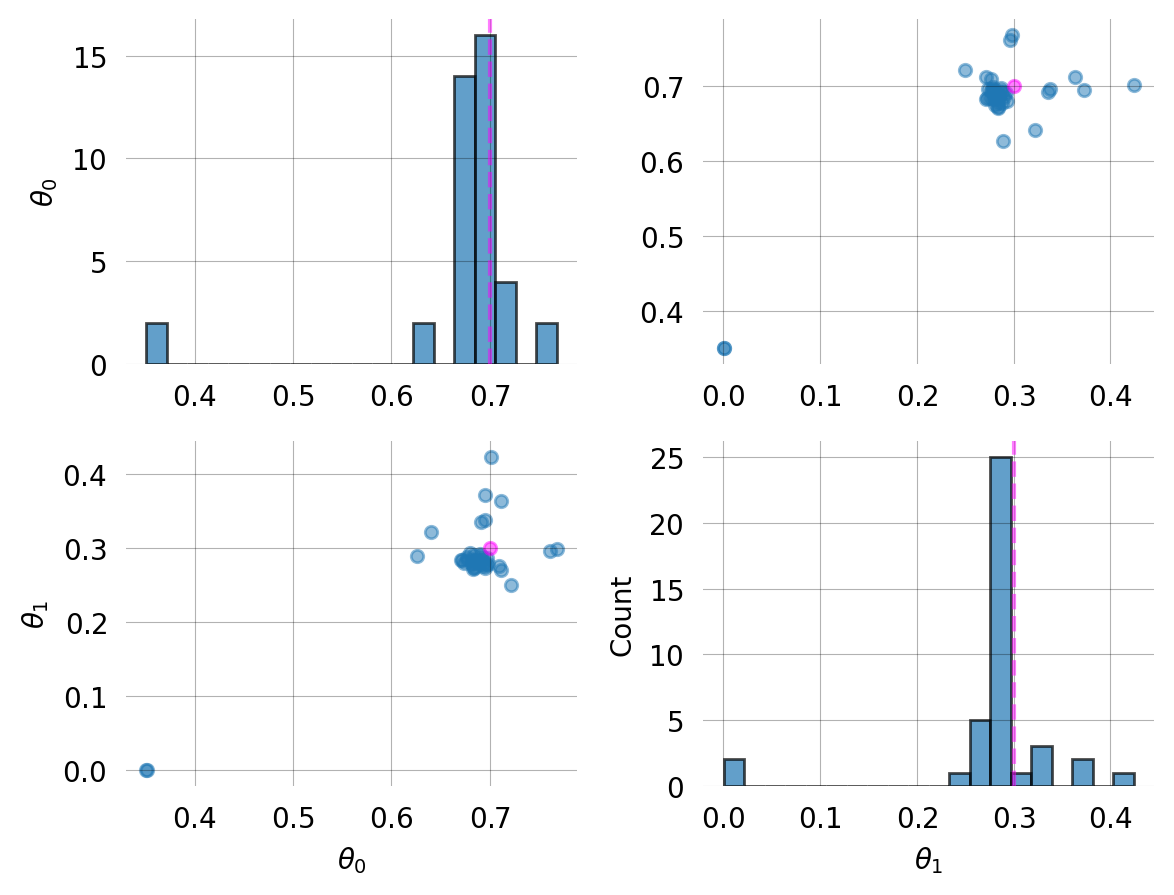
svgd = graph.svgd(
observed_data,
prior=mom.prior,
optimizer=Adam(0.25),
)
svgd.summary()
svgd.plot_convergence() ;Parameter Fixed MAP Mean SD HPD 95% lo HPD 95% hi
0 No 0.6944 0.6742 0.0777 0.6259 0.7680
1 No 0.2802 0.2794 0.0714 0.2500 0.4239
Particles: 40, Iterations: 100Fixed parameters
When some parameters are known, you can fix them during moment matching. The fixed argument takes a list of (index, value) tuples. Only the free parameters are optimised, and the returned prior list contains None at the fixed positions.
Here we fix the coalescent rate and estimate only the migration rate:
mom_fixed = graph.method_of_moments(
observed_data,
fixed=[(0, 0.7)], # fix theta[0] (coalescent rate) at 0.7
)
print(f"MoM estimate: {mom_fixed.theta}")
print(f"Priors: {mom_fixed.prior}")
print(f" theta[0] prior is None (fixed): {mom_fixed.prior[0] is None}")
print(f" theta[1] prior: mean={mom_fixed.prior[1].mu:.3f}, std={mom_fixed.prior[1].sigma:.3f}")MoM: theta_dim=2, n_free=1, nr_moments=4, n_features=1, n_equations=4
MoM: sample moments =
[2.89041844e+00 2.19034775e+01 2.64810856e+02 4.27038260e+03]
MoM: initial guess (full theta) = [0.7 8.21283765]
MoM: converged — Both `ftol` and `xtol` termination conditions are satisfied.
MoM: theta = [0.7 0.26600949]
MoM: residual = 6.416370e+00
MoM: model moments =
[2.85714286e+00 2.16969082e+01 2.62288310e+02 4.27047933e+03]
MoM estimate: [0.7 0.26600949]
Priors: [None, <phasic.svgd.GaussPrior object at 0x360b132d0>]
theta[0] prior is None (fixed): True
theta[1] prior: mean=0.266, std=0.132The fixed parameter priors integrate seamlessly with SVGD’s fixed argument:
svgd_fixed = graph.svgd(
observed_data,
prior=mom_fixed.prior,
fixed=[(0, 0.7)],
optimizer=Adam(0.25),
)
svgd_fixed.summary()Parameter Fixed MAP Mean SD HPD 95% lo HPD 95% hi
0 Yes 0.7000 NA NA NA NA
1 No 0.2803 0.2828 0.0310 0.2392 0.3174
Particles: 40, Iterations: 100Multi-feature observations with rewards
For models where observations are reward-transformed (e.g. branch lengths contributing to different mutation categories in population genetics), method of moments can match moments per feature. Each feature has its own reward vector, and the combined system of equations constrains the parameters.
graph_1p = Graph(coalescent_1param)
true_theta_1p = [7]
graph_1p.update_weights(true_theta_1p)
# Create reward vectors — each row is a feature's reward across all vertices
states = graph_1p.states().T
rewards = states[:-1] # one row per feature (e.g. singleton, doubleton, tripleton branch lengths)
print(f"Reward matrix shape: {rewards.shape} (n_features={rewards.shape[0]}, n_vertices={rewards.shape[1]})")
print(f"Rewards:\n{rewards}")Reward matrix shape: (3, 6) (n_features=3, n_vertices=6)
Rewards:
[[0 4 2 0 1 0]
[0 0 1 2 0 0]
[0 0 0 0 1 0]]# Sample multi-feature observations (each SNP contributes to one feature)
n_obs = 10000
n_features = rewards.shape[0]
observed_data_2d = np.zeros((n_obs * n_features, n_features), dtype=float)
observed_data_2d[:] = np.nan
for i in range(n_features):
observed_data_2d[(i*n_obs):((i+1)*n_obs), i] = graph_1p.sample(n_obs, rewards=rewards[i])
np.random.shuffle(observed_data_2d)
print(f"Observation shape: {observed_data_2d.shape}")
print(f"First 5 rows (NaN = unobserved feature):")
print(observed_data_2d[:5])Observation shape: (30000, 3)
First 5 rows (NaN = unobserved feature):
[[0.12124815 nan nan]
[0.36418885 nan nan]
[ nan nan 0.10161367]
[0.03089352 nan nan]
[0.60967296 nan nan]]mom_multi = graph_1p.method_of_moments(
observed_data_2d,
rewards=rewards,
)
print(f"True theta: {true_theta_1p}")
print(f"MoM estimate: {mom_multi.theta}")
print(f"Converged: {mom_multi.success}")
print(f"\nSample moments (n_features x nr_moments):\n{mom_multi.sample_moments}")
print(f"\nModel moments (n_features x nr_moments):\n{mom_multi.model_moments}")MoM: theta_dim=1, n_free=1, nr_moments=4, n_features=3, n_equations=12
MoM: sample moments =
[[0.28569414 0.11867841 0.06469594 0.04371687]
[0.1425845 0.06761152 0.05566143 0.06176143]
[0.09469394 0.02795647 0.01293379 0.00821446]]
MoM: initial guess (full theta) = [6.95192796]
MoM: converged — `gtol` termination condition is satisfied.
MoM: theta = [7.00133145]
MoM: residual = 1.515105e-05
MoM: model moments =
[[0.28565995 0.11786899 0.06345585 0.04217254]
[0.14282998 0.06800134 0.05633325 0.06399868]
[0.09521998 0.02720054 0.01165516 0.00665882]]
True theta: [7]
MoM estimate: [7.00133145]
Converged: True
Sample moments (n_features x nr_moments):
[[0.28569414 0.11867841 0.06469594 0.04371687]
[0.1425845 0.06761152 0.05566143 0.06176143]
[0.09469394 0.02795647 0.01293379 0.00821446]]
Model moments (n_features x nr_moments):
[[0.28565995 0.11786899 0.06345585 0.04217254]
[0.14282998 0.06800134 0.05633325 0.06399868]
[0.09521998 0.02720054 0.01165516 0.00665882]]The multivariate MoM priors can be used with multivariate SVGD:
FIXME:
svgd_multi = graph_1p.svgd( observed_data_2d, rewards=rewards, prior=mom_multi.prior, optimizer=Adam(0.25), ) svgd_multi.summary() svgd_multi.plot_convergence() ;
Controlling the number of moments
By default, method_of_moments automatically selects the number of moments to create an overdetermined system: max(2 × n_free_params, 4) moments per feature. This typically gives robust estimates without any tuning.
You can override the automatic selection by passing nr_moments explicitly. If you specify fewer moment equations than free parameters, the method automatically increases nr_moments:
FIXME:
graph_2p = Graph(coalescent_islands) graph_2p.update_weights([0.7, 0.3]) data_2p = graph_2p.sample(1000)
Ask for 1 moment with 2 free params -> auto-increases
mom_auto = graph_2p.method_of_moments(data_2p, nr_moments=1)
Using more moments than parameters (overdetermined system) can improve robustness:
mom_over = graph_2p.method_of_moments(data_2p, nr_moments=4) print(f”nr_moments=4, theta = {mom_over.theta}“) print(f”Residual: {mom_over.residual:.2e}“)
Adjusting the prior width
The std_multiplier parameter controls how wide the MoM-derived prior is relative to the asymptotic standard error. A larger multiplier gives a wider, more permissive prior:
# Default: std_multiplier=2.0
mom_default = graph.method_of_moments(observed_data, verbose=False)
print(f"std_multiplier=2.0 -> prior std = {mom_default.prior[0].sigma:.4f}")
# Wider prior
mom_wide = graph.method_of_moments(observed_data, std_multiplier=5.0, verbose=False)
print(f"std_multiplier=5.0 -> prior std = {mom_wide.prior[0].sigma:.4f}")
# Tighter prior
mom_tight = graph.method_of_moments(observed_data, std_multiplier=1.0, verbose=False)
print(f"std_multiplier=1.0 -> prior std = {mom_tight.prior[0].sigma:.4f}")std_multiplier=2.0 -> prior std = 0.0872
std_multiplier=5.0 -> prior std = 0.2179
std_multiplier=1.0 -> prior std = 0.0436The MoMResult object
The method_of_moments method returns a MoMResult dataclass with the following fields:
| Field | Type | Description |
|---|---|---|
theta |
np.ndarray |
MoM parameter estimate |
std |
np.ndarray |
Standard error of the MoM estimator (delta method) |
prior |
list |
List of GaussPrior objects (or None for fixed params) |
success |
bool |
Whether the optimisation converged |
residual |
float |
Sum of squared residuals |
sample_moments |
np.ndarray |
Empirical moments from the data |
model_moments |
np.ndarray |
Model moments at the solution |
message |
str |
Solver status message |
The standard errors quantify the sampling uncertainty of the MoM estimator — how much theta would change with a different random sample of the same size. They are computed via the delta method, propagating the covariance of the sample moments through the moment-matching equations. These errors scale as 1/\sqrt{n} with the sample size n.
Complete workflow
Putting it all together, the recommended workflow for inference is:
- Build the parameterized graph
- MoM — get data-informed priors in seconds
- SVGD — run full Bayesian inference with the MoM priors
- Diagnose — check convergence and posterior
# 1. Build model
graph = Graph(coalescent_islands)
# 2. Method of moments
true_theta = [0.7, 0.3]
graph.update_weights(true_theta)
observations = graph.sample(100)
mom = graph.method_of_moments(observations)MoM: theta_dim=2, n_free=2, nr_moments=4, n_features=1, n_equations=4
MoM: sample moments =
[ 2.42023367 14.23982075 127.14644098 1490.61257429]
MoM: initial guess (full theta) = [0.910071 7.07235481]
MoM: converged — Both `ftol` and `xtol` termination conditions are satisfied.
MoM: theta = [0.79598546 0.6802256 ]
MoM: residual = 6.607437e-02
MoM: model moments =
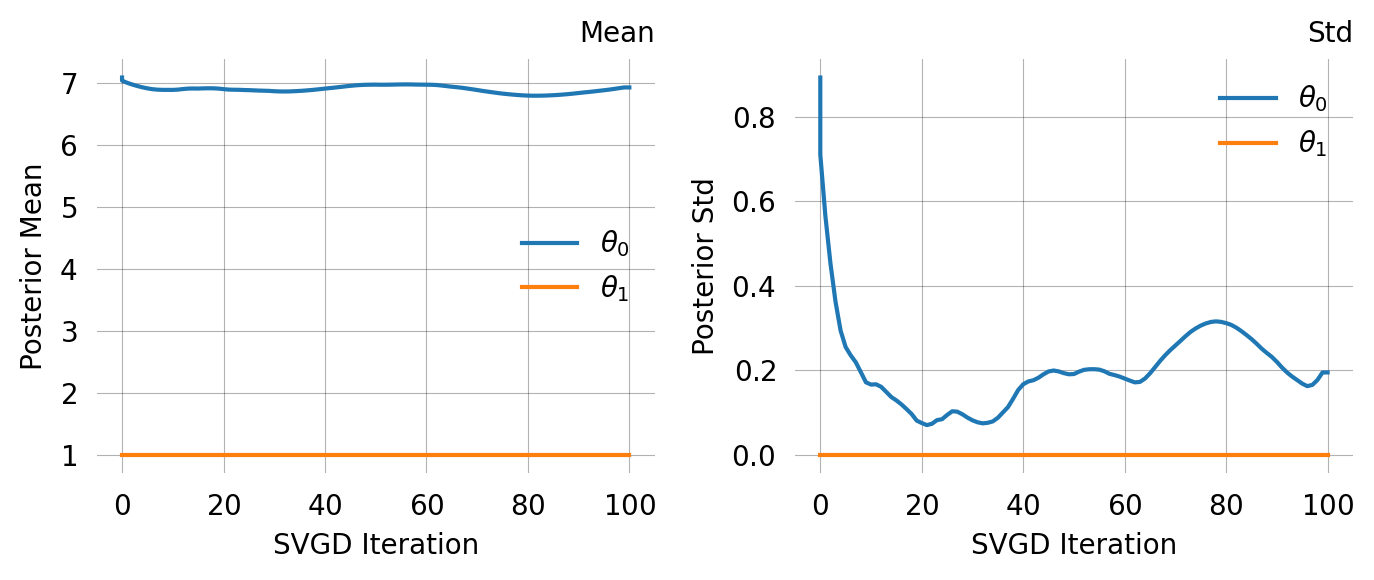
[ 2.51260871 14.47329878 127.09145244 1490.61491816]fig, axes = plt.subplots(len(mom.prior), 1, figsize=(6,4))
for i, prior in enumerate(mom.prior):
prior.plot(return_ax=True, ax=axes[i])
plt.tight_layout()# from phasic import configure
# configure(
# force_high_precision=True,
# mpfr_precision_bits=256
# )
# svgd = graph.svgd(
# observations,
# prior=mom.prior,
# learning_rate=ExpStepSize(first_step=0.01, last_step=0.001, tau=20.0),
# n_iterations=100,
# )# svgd.summary()# svgd.plot_convergence()# svgd.plot_trace()# svgd.plot_pairwise(true_theta=true_theta)# svgd.plot_hdr()Joint probability models
For joint probability graphs — created via graph.joint_prob_graph() — observations are feature-count tuples (e.g., [0, 1, 0, 0]) rather than continuous times. The model outputs a probability table, not a PDF/PMF with moments, so standard moment matching does not apply.
The probability_matching() method handles this case by matching the empirical probability distribution to the model probability distribution:
\hat{\theta}_{\text{PM}} = \arg\min_{\theta > 0} \left\| \mathbf{p}_{\text{model}}(\theta) - \hat{\mathbf{p}} \right\|^2
where \hat{\mathbf{p}} are the empirical proportions of each observation pattern. Standard errors are obtained via the delta method with the multinomial covariance:
\text{Cov}(\hat{p}_i, \hat{p}_j) = \frac{\delta_{ij} p_i - p_i p_j}{n}
from functools import partial
from itertools import combinations_with_replacement
all_pairs = partial(combinations_with_replacement, r=2)
# Build the coalescent base graph
nr_samples = 3
indexer = StateIndexer(
lineage=[
Property('descendants', min_value=1, max_value=nr_samples),
]
)
@with_ipv([nr_samples]+[0]*(nr_samples-1))
def coalescent_1param(state):
transitions = []
for i, j in all_pairs(indexer.lineage):
p1 = indexer.lineage.index_to_props(i)
p2 = indexer.lineage.index_to_props(j)
same = int(i == j)
if same and state[i] < 2:
continue
if not same and (state[i] < 1 or state[j] < 1):
continue
new = state.copy()
new[i] -= 1
new[j] -= 1
descendants = p1.descendants + p2.descendants
k = indexer.lineage.props_to_index(descendants=descendants)
new[k] += 1
transitions.append([new, [state[i]*(state[j]-same)/(1+same)]])
return transitions
base_graph = Graph(coalescent_1param)
# Create joint probability graph
mutation_rate = 1.0
joint_graph = base_graph.joint_prob_graph(
indexer, tot_reward_limit=2, mutation_rate=mutation_rate
)
joint_graph.joint_prob_table()| descendants_1 | descendants_2 | descendants_3 | prob | |
|---|---|---|---|---|
| t_vertex_index | ||||
| 4 | 0 | 0 | 0 | 0.166667 |
| 8 | 1 | 0 | 0 | 0.138889 |
| 11 | 0 | 1 | 0 | 0.055556 |
| 13 | 2 | 0 | 0 | 0.043981 |
| 14 | 1 | 1 | 0 | 0.032407 |
| 15 | 0 | 2 | 0 | 0.009259 |
Sample observations from the joint probability table:
true_theta = [7.0, mutation_rate]
joint_graph.update_weights(true_theta)
# Sample from the joint probability table
table = joint_graph.joint_prob_table()
p = table['prob'].to_numpy()
p = p / p.sum()
feature_cols = table.columns[:-1]
np.random.seed(42)
n_obs = 2000
sampled_rows = np.random.choice(len(table), size=n_obs, p=p)
observations = [tuple(int(x) for x in row) for row in table.iloc[sampled_rows][feature_cols].values]
print(f"Sampled {n_obs} observations, first 5: {observations[:5]}")Sampled 2000 observations, first 5: [(0, 0, 0), (2, 0, 0), (1, 0, 0), (0, 0, 0), (0, 0, 0)]Run probability_matching() with the mutation rate fixed:
pm = joint_graph.probability_matching(
observations,
fixed=[(1, mutation_rate)],
)
print(f"True theta: {true_theta}")
print(f"PM estimate: {pm.theta}")
print(f"Std error: {pm.std}")
print(f"Converged: {pm.success}")
print(f"Residual: {pm.residual:.2e}")ProbMatch: theta_dim=2, n_free=1, n_obs=2000, n_unique_obs=6
ProbMatch: initial guess (full theta) = [7.88046282 1. ]
ProbMatch: converged — `gtol` termination condition is satisfied.
ProbMatch: theta = [7.00204262 1. ]
ProbMatch: residual = 3.839769e-05
True theta: [7.0, 1.0]
PM estimate: [7.00204262 1. ]
Std error: [0.35605011 0. ]
Converged: True
Residual: 3.84e-05The ProbMatchResult includes the empirical and model probabilities at the solution:
import pandas as pd
pd.DataFrame({
'vertex_index': pm.unique_indices,
'empirical_prob': pm.empirical_probs,
'model_prob': pm.model_probs,
})| vertex_index | empirical_prob | model_prob | |
|---|---|---|---|
| 0 | 4 | 0.6935 | 0.694502 |
| 1 | 8 | 0.1590 | 0.163940 |
| 2 | 11 | 0.0785 | 0.077149 |
| 3 | 13 | 0.0310 | 0.029057 |
| 4 | 14 | 0.0295 | 0.026782 |
| 5 | 15 | 0.0085 | 0.008570 |
The .prior list can be passed directly to graph.svgd(), just like MoMResult.prior:
svgd_jp = joint_graph.svgd(
observations,
prior=pm.prior,
fixed=[(1, mutation_rate)],
optimizer=Adam(0.25),
)svgd_jp.summary()Parameter Fixed MAP Mean SD HPD 95% lo HPD 95% hi
0 No 6.8955 6.9337 0.1944 6.5354 7.2389
1 Yes 1.0000 NA NA NA NA
Particles: 40, Iterations: 100One-step alternative: DataPrior
The two-step pattern of running method_of_moments (or probability_matching) followed by passing .prior to graph.svgd() is common enough that DataPrior wraps it into a single object. It auto-detects whether the graph is a joint probability graph and calls the appropriate estimation method.
# Before (two steps):
mom = graph.method_of_moments(data, fixed=[(1, 1.0)])
svgd = graph.svgd(data, prior=mom.prior, fixed=[(1, 1.0)])
# After (one step):
svgd = graph.svgd(data, prior=DataPrior(graph, data, fixed=[(1, 1.0)]), fixed=[(1, 1.0)])# from phasic import DataPrior
# # Standard graph — DataPrior calls method_of_moments internally
# graph_dp = Graph(coalescent_islands)
# graph_dp.update_weights([0.7, 0.3])
# data_dp = graph_dp.sample(500)
# dp = DataPrior(graph_dp, data_dp, sd=2.0)
# print(dp)
# print(f"Underlying method: {dp.method}")
# print(f"Theta estimate: {dp.theta}")
# print(f"Converged: {dp.success}")
# # Use directly as the prior argument
# svgd_dp = graph_dp.svgd(data_dp, prior=dp, n_iterations=50, verbose=False)
# svgd_dp.summary()# # Joint probability graph — DataPrior calls probability_matching internally
# dp_joint = DataPrior(
# joint_graph, observations,
# fixed=[(1, mutation_rate)],
# )
# print(dp_joint)
# print(f"Underlying method: {dp_joint.method}")